Note
To run this notebook in JupyterLab, load examples/ex7_1.ipynb
Statistical Relational Learning with pslpython¶
In this section we'll explore one form of statistical relational learning called probabilistic soft logic (PSL).
One of the examples given for PSL is called simple acquaintances, which uses a graph of some friends, where they live, what interests they share, and then infers who probably knows whom. Some people explicitly do or do not know each other, while other "knows" relations can be inferred based on whether two people have lived in the same place or share common interest.
The objective is to build a PSL model for link prediction, to evaluate the annotations in the friend graph. In this case, we'll assume that the "knows" relations have been added from a questionable source (e.g., some third-party dataset) so we'll measure a subset of these relations and determine their likelihood. NB: this is really useful for cleaning up annotations in a large graph!
Now let's load a KG which is an RDF representation of this "simple acquaintances" example, based on using the foaf vocabulary:
from os.path import dirname
import kglab
import os
namespaces = {
"acq": "http://example.org/stuff/",
"foaf": "http://xmlns.com/foaf/0.1/",
}
kg = kglab.KnowledgeGraph(
name = "LINQS simple acquaintance example for PSL",
base_uri = "http://example.org/stuff/",
namespaces = namespaces,
)
kg.load_rdf(dirname(os.getcwd()) + "/dat/acq.ttl") ;
Take a look at the dat/acq.ttl file to see the people and their relations.
Here's a quick visualization of the graph:
VIS_STYLE = {
"foaf": {
"color": "orange",
"size": 5,
},
"acq":{
"color": "blue",
"size": 30,
},
}
excludes = [
kg.get_ns("rdf").type,
kg.get_ns("rdfs").domain,
kg.get_ns("rdfs").range,
]
subgraph = kglab.SubgraphTensor(kg, excludes=excludes)
pyvis_graph = subgraph.build_pyvis_graph(notebook=True, style=VIS_STYLE)
pyvis_graph.force_atlas_2based()
pyvis_graph.show("tmp.fig04.html")
Loading a PSL model¶
Next, we'll use the pslpython library implemented in Python (atop its core library running in Java) to define three predicates (i.e., relations – similar as in RDF) which are: Neighbors, Likes, Knows
psl = kglab.PSLModel(
name = "simple acquaintances",
)
Then add each of the predicates:
psl.add_predicate("Lived", size=2)
psl.add_predicate("Likes", size=2)
psl.add_predicate("Knows", size=2, closed=False)
;
''
Next, we'll add a set of probabilistic rules, all with different weights applied:
- "Two people who live in the same place are more likely to know each other"
- "Two people who don't live in the same place are less likely to know each other"
- "Two people who share a common interest are more likely to know each other"
- "Two people who both know a third person are more likely to know each other"
- "Otherwise, any pair of people are less likely to know each other"
psl.add_rule("Lived(P1, L) & Lived(P2, L) & (P1 != P2) -> Knows(P1, P2)", weight=20.0, squared=True)
psl.add_rule("Lived(P1, L1) & Lived(P2, L2) & (P1 != P2) & (L1 != L2) -> !Knows(P1, P2)", weight=5.0, squared=True)
psl.add_rule("Likes(P1, L) & Likes(P2, L) & (P1 != P2) -> Knows(P1, P2)", weight=10.0, squared=True)
psl.add_rule("Knows(P1, P2) & Knows(P2, P3) & (P1 != P3) -> Knows(P1, P3)", weight=5.0, squared=True)
psl.add_rule("!Knows(P1, P2)", weight=5.0, squared=True)
;
''
Finally we'll add a commutative rule such that:
"If Person 1 knows Person 2, then Person 2 also knows Person 1."
psl.add_rule("Knows(P1, P2) = Knows(P2, P1)", weighted=False) ;
To initialize the model, we'll clear any pre-existing data for each of the predicates:
psl.clear_model() ;
Next we'll create a specific Subgraph to transform the names of foaf:Person in the graph, since the PSL rules in this example focus on relations among the people:
people_iter = kg.rdf_graph().subjects(kg.get_ns("rdf").type, kg.get_ns("foaf").Person)
people_nodes = [ p for p in sorted(people_iter, key=lambda p: str(p)) ]
subgraph_people = kglab.Subgraph(kg, preload=people_nodes)
Now let's query our KG to populate data into the Liked predicate in the PSL model, based on foaf:based_near which represents people who live nearby each other:
sparql = """
SELECT DISTINCT ?p1 ?l
WHERE {
?p1 foaf:based_near ?l
}
"""
for row in kg.query(sparql):
p1 = subgraph_people.transform(row.p1)
l = subgraph.transform(row.l)
psl.add_data_row("Lived", [p1, l])
Note: these data points are observations, i.e., empirical support for the probabilistic model.
Next let's query our KG to populate data into the Likes predicate in the PSL model, based on shared interests in foaf:topic_interest topics:
sparql = """
SELECT DISTINCT ?p1 ?t
WHERE {
?p1 foaf:topic_interest ?t
}
"""
for row in kg.query(sparql):
p1 = subgraph_people.transform(row.p1)
t = subgraph.transform(row.t)
psl.add_data_row("Likes", [p1, t])
Just for kicks, let's take a look at the internal representation of a PSL predicate, which is a pandas.DataFrame:
predicate = psl.model.get_predicate("Likes")
predicate.__dict__
{'_types': [<ArgType.UNIQUE_STRING_ID: 'UniqueStringID'>,
<ArgType.UNIQUE_STRING_ID: 'UniqueStringID'>],
'_data': {<Partition.OBSERVATIONS: 'observations'>: 0 1 2
0 0 70 1.0
1 1 70 1.0
2 2 70 1.0
3 3 70 1.0
4 4 70 1.0
.. .. .. ...
127 17 79 1.0
128 18 79 1.0
129 21 79 1.0
130 22 79 1.0
131 24 79 1.0
[132 rows x 3 columns],
<Partition.TARGETS: 'targets'>: Empty DataFrame
Columns: [0, 1, 2]
Index: [],
<Partition.TRUTH: 'truth'>: Empty DataFrame
Columns: [0, 1, 2]
Index: []},
'_name': 'LIKES',
'_closed': False}
df = psl.trace_predicate("Likes", partition="observations")
df
| P1 | P2 | value | |
|---|---|---|---|
| 0 | 0 | 70 | 1.0 |
| 1 | 0 | 71 | 1.0 |
| 2 | 0 | 72 | 1.0 |
| 3 | 0 | 73 | 1.0 |
| 4 | 0 | 75 | 1.0 |
| ... | ... | ... | ... |
| 127 | 24 | 73 | 1.0 |
| 128 | 24 | 74 | 1.0 |
| 129 | 24 | 75 | 1.0 |
| 130 | 24 | 78 | 1.0 |
| 131 | 24 | 79 | 1.0 |
132 rows × 3 columns
Now we'll load data from the dat/psl/knows_targets.txt CSV file, which is a list of foaf:knows relations in our graph that we want to analyze.
Each of these has an assumed value of 1.0 (true) or 0.0 (false).
Our PSL analysis will assign probabilities for each so that we can compare which annotations appear to be suspect and require further review:
import csv
import pandas as pd
targets = []
rows_list = []
with open(dirname(os.getcwd()) + "/dat/psl/knows_targets.txt", "r") as f:
reader = csv.reader(f, delimiter="\t")
for i, row in enumerate(reader):
p1 = int(row[0])
p2 = int(row[1])
targets.append((p1, p2))
p1_node = subgraph_people.inverse_transform(p1)
p2_node = subgraph_people.inverse_transform(p2)
if (p1_node, kg.get_ns("foaf").knows, p2_node) in kg.rdf_graph():
truth = 1.0
rows_list.append({ 0: p1, 1: p2, "truth": truth})
psl.add_data_row("Knows", [p1, p2], partition="truth", truth_value=truth)
psl.add_data_row("Knows", [p1, p2], partition="targets")
elif (p1_node, kg.get_ns("acq").wantsIntro, p2_node) in kg.rdf_graph():
truth = 0.0
rows_list.append({ 0: p1, 1: p2, "truth": truth})
psl.add_data_row("Knows", [p1, p2], partition="truth", truth_value=truth)
psl.add_data_row("Knows", [p1, p2], partition="targets")
else:
print("UNKNOWN", p1, p2)
Here are data points which are considered ground atoms, each with a truth value set initially.
Here are also our targets for which nodes in the graph to analyze based on the rules.
We'll keep a dataframe called df_dat to preserve these values for later use:
df_dat = pd.DataFrame(rows_list)
df_dat.head()
| 0 | 1 | truth | |
|---|---|---|---|
| 0 | 0 | 1 | 1.0 |
| 1 | 0 | 7 | 1.0 |
| 2 | 0 | 15 | 1.0 |
| 3 | 0 | 18 | 1.0 |
| 4 | 0 | 22 | 0.0 |
Next, we'll add foaf:knows observations which are in the graph, although not among our set of targets.
This provides more evidence for the probabilistic inference.
Note that since RDF does not allow for representing probabilities on relations, we're using the acq:wantsIntro to represent a foaf:knows with a 0.0 probability:
sparql = """
SELECT ?p1 ?p2
WHERE {
?p1 foaf:knows ?p2 .
}
ORDER BY ?p1 ?p2
"""
for row in kg.query(sparql):
p1 = subgraph_people.transform(row.p1)
p2 = subgraph_people.transform(row.p2)
if (p1, p2) not in targets:
psl.add_data_row("Knows", [p1, p2], truth_value=1.0)
sparql = """
SELECT ?p1 ?p2
WHERE {
?p1 acq:wantsIntro ?p2 .
}
ORDER BY ?p1 ?p2
"""
for row in kg.query(sparql):
p1 = subgraph_people.transform(row.p1)
p2 = subgraph_people.transform(row.p2)
if (p1, p2) not in targets:
psl.add_data_row("Knows", [p1, p2], truth_value=0.0)
Now we're ready to run optimization on the PSL model and infer the grounded atoms. This may take a few minutes to run:
psl.infer()
6955 [pslpython.model PSL] INFO --- 0 [main] INFO org.linqs.psl.cli.Launcher - Running PSL CLI Version 2.2.2-5f9a472
7289 [pslpython.model PSL] INFO --- 335 [main] INFO org.linqs.psl.cli.Launcher - Loading data
7536 [pslpython.model PSL] INFO --- 583 [main] INFO org.linqs.psl.cli.Launcher - Data loading complete
7537 [pslpython.model PSL] INFO --- 583 [main] INFO org.linqs.psl.cli.Launcher - Loading model from /var/folders/dp/q971mmvs2m98ypxb3sb0xmxc0000gn/T/psl-python/simple acquaintances/simple acquaintances.psl
7633 [pslpython.model PSL] INFO --- 679 [main] INFO org.linqs.psl.cli.Launcher - Model loading complete
7633 [pslpython.model PSL] INFO --- 680 [main] INFO org.linqs.psl.cli.Launcher - Starting inference with class: org.linqs.psl.application.inference.MPEInference
7716 [pslpython.model PSL] INFO --- 763 [main] INFO org.linqs.psl.application.inference.MPEInference - Grounding out model.
7901 [pslpython.model PSL] INFO --- 948 [main] INFO org.linqs.psl.application.inference.MPEInference - Grounding complete.
7923 [pslpython.model PSL] INFO --- 970 [main] INFO org.linqs.psl.application.inference.InferenceApplication - Beginning inference.
8715 [pslpython.model PSL] WARNING --- 1762 [main] WARN org.linqs.psl.reasoner.admm.ADMMReasoner - No feasible solution found. 34 constraints violated.
8716 [pslpython.model PSL] INFO --- 1762 [main] INFO org.linqs.psl.reasoner.admm.ADMMReasoner - Optimization completed in 1009 iterations. Objective: 7542.3315, Feasible: false, Primal res.: 0.053818043, Dual res.: 0.0010119631
8716 [pslpython.model PSL] INFO --- 1762 [main] INFO org.linqs.psl.application.inference.InferenceApplication - Inference complete.
8717 [pslpython.model PSL] INFO --- 1762 [main] INFO org.linqs.psl.application.inference.InferenceApplication - Writing results to Database.
8748 [pslpython.model PSL] INFO --- 1794 [main] INFO org.linqs.psl.application.inference.InferenceApplication - Results committed to database.
8749 [pslpython.model PSL] INFO --- 1794 [main] INFO org.linqs.psl.cli.Launcher - Inference Complete
Let's examine the results.
We'll get a pandas.DataFrame describing the targets in the Knows predicate:
df = psl.get_results("Knows")
df.head()
| predicate | 0 | 1 | truth | |
|---|---|---|---|---|
| 0 | KNOWS | 16 | 15 | 0.998196 |
| 1 | KNOWS | 16 | 23 | 0.000551 |
| 2 | KNOWS | 17 | 18 | 0.561153 |
| 3 | KNOWS | 17 | 22 | 0.994712 |
| 4 | KNOWS | 17 | 21 | 0.476654 |
Now we can compare the "truth" values from our targets, with their probabilities from the inference provided by the PSL model. Let's build a dataframe to show that:
dat_val = {}
df.insert(1, "p1", "")
df.insert(2, "p2", "")
for index, row in df_dat.iterrows():
p1 = int(row[0])
p2 = int(row[1])
key = (p1, p2)
dat_val[key] = row["truth"]
for index, row in df.iterrows():
p1 = int(row[0])
p2 = int(row[1])
key = (p1, p2)
df.at[index, "diff"] = row["truth"] - dat_val[key]
df.at[index, "p1"] = str(subgraph_people.inverse_transform(p1))
df.at[index, "p2"] = str(subgraph_people.inverse_transform(p2))
df = df.drop(df.columns[[3, 4]], axis=1)
df.head()
| predicate | p1 | p2 | truth | diff | |
|---|---|---|---|---|---|
| 0 | KNOWS | http://example.org/stuff/person_16 | http://example.org/stuff/person_15 | 0.998196 | -0.001804 |
| 1 | KNOWS | http://example.org/stuff/person_16 | http://example.org/stuff/person_23 | 0.000551 | 0.000551 |
| 2 | KNOWS | http://example.org/stuff/person_17 | http://example.org/stuff/person_18 | 0.561153 | 0.561153 |
| 3 | KNOWS | http://example.org/stuff/person_17 | http://example.org/stuff/person_22 | 0.994712 | -0.005288 |
| 4 | KNOWS | http://example.org/stuff/person_17 | http://example.org/stuff/person_21 | 0.476654 | 0.476654 |
In other words, which of these "knows" relations in the graph appears to be suspect, based on our rules plus the other evidence in the graph?
Let's visualize a histogram of how the inferred probabilities are distributed:
df["diff"].hist();
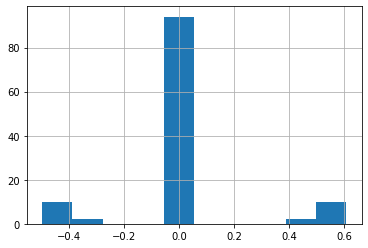
In most cases there is little or no difference in the probabilities for the target relations.
However, some appear to be off by a substantial (0.6) amount, which indicates potential problems in this part of our graph data.
The following rows show where these foaf:knows annotations in the graph differs significantly from their truth values predicted by PSL:
df[df["diff"] >= 0.4]
| predicate | p1 | p2 | truth | diff | |
|---|---|---|---|---|---|
| 2 | KNOWS | http://example.org/stuff/person_17 | http://example.org/stuff/person_18 | 0.561153 | 0.561153 |
| 4 | KNOWS | http://example.org/stuff/person_17 | http://example.org/stuff/person_21 | 0.476654 | 0.476654 |
| 5 | KNOWS | http://example.org/stuff/person_18 | http://example.org/stuff/person_17 | 0.560540 | 0.560540 |
| 15 | KNOWS | http://example.org/stuff/person_14 | http://example.org/stuff/person_06 | 0.507943 | 0.507943 |
| 24 | KNOWS | http://example.org/stuff/person_19 | http://example.org/stuff/person_09 | 0.539348 | 0.539348 |
| 28 | KNOWS | http://example.org/stuff/person_17 | http://example.org/stuff/person_01 | 0.581051 | 0.581051 |
| 36 | KNOWS | http://example.org/stuff/person_01 | http://example.org/stuff/person_17 | 0.578490 | 0.578490 |
| 39 | KNOWS | http://example.org/stuff/person_21 | http://example.org/stuff/person_17 | 0.476276 | 0.476276 |
| 55 | KNOWS | http://example.org/stuff/person_09 | http://example.org/stuff/person_19 | 0.540269 | 0.540269 |
| 70 | KNOWS | http://example.org/stuff/person_06 | http://example.org/stuff/person_14 | 0.506647 | 0.506647 |
| 84 | KNOWS | http://example.org/stuff/person_06 | http://example.org/stuff/person_09 | 0.606492 | 0.606492 |
| 91 | KNOWS | http://example.org/stuff/person_09 | http://example.org/stuff/person_06 | 0.607750 | 0.607750 |
In most of these cases, the truth value is floating (somewhere near ~0.5) when it was expected to be zero (i.e., they don't know each other).
Part of that likely comes from the use of the Likes predicate with boolean values; the original demo had probabilities for those, but was simplified here.
Speaking of human-in-the-loop practices for AI, using PSL along with a KG seems like a great way to leverage machine learning, so that the people can focus on parts of the graph that have the most uncertainty. And, therefore, probably provide the best ROI for investing time+cost into curation.
Exercises¶
Exercise 1:
Build a PSL model that tests the "noodle vs. pancake" rules used in an earlier example with our recipe KG. Which recipes should be annotated differently?
Exercise 2:
Try representing one of the other PSL examples using RDF and kglab.